Building Cython code¶
Cython code must, unlike Python, be compiled. This happens in two stages:
- A .pyx file is compiled by Cython to a .c file, containing the code of a Python extension module
- The .c file is compiled by a C compiler to a .so file (or .pyd on Windows) which can be import-ed directly into a Python session.
There are several ways to build Cython code:
- Write a distutils setup.py.
- Use pyximport, importing Cython .pyx files as if they were .py files (using distutils to compile and build in the background).
- Run the cython command-line utility manually to produce the .c file from the .pyx file, then manually compiling the .c file into a shared object library or DLL suitable for import from Python. (These manual steps are mostly for debugging and experimentation.)
- Use the [IPython] notebook or the [Sage] notebook, both of which allow Cython code inline.
Currently, distutils is the most common way Cython files are built and distributed. The other methods are described in more detail in the Source Files and Compilation section of the reference manual.
Building a Cython module using distutils¶
Imagine a simple “hello world” script in a file hello.pyx:
def say_hello_to(name):
print("Hello %s!" % name)
The following could be a corresponding setup.py script:
from distutils.core import setup
from Cython.Build import cythonize
setup(
name = 'Hello world app',
ext_modules = cythonize("hello.pyx"),
)
To build, run python setup.py build_ext --inplace. Then simply start a Python session and do from hello import say_hello_to and use the imported function as you see fit.
Using the IPython notebook¶
Cython can be used conveniently and interactively from a web browser through the IPython notebook. To install IPython, e.g. into a virtualenv, use pip:
(venv)$ pip install "ipython[notebook]"
(venv)$ ipython notebook
To enable support for Cython compilation, install Cython and load the cythonmagic extension from within IPython:
%load_ext cythonmagic
Then, prefix a cell with the %%cython marker to compile it:
%%cython
cdef int a = 0
for i in range(10):
a += i
print a
You can show Cython’s code analysis by passing the --annotate option:
%%cython --annotate
...
Using the Sage notebook¶
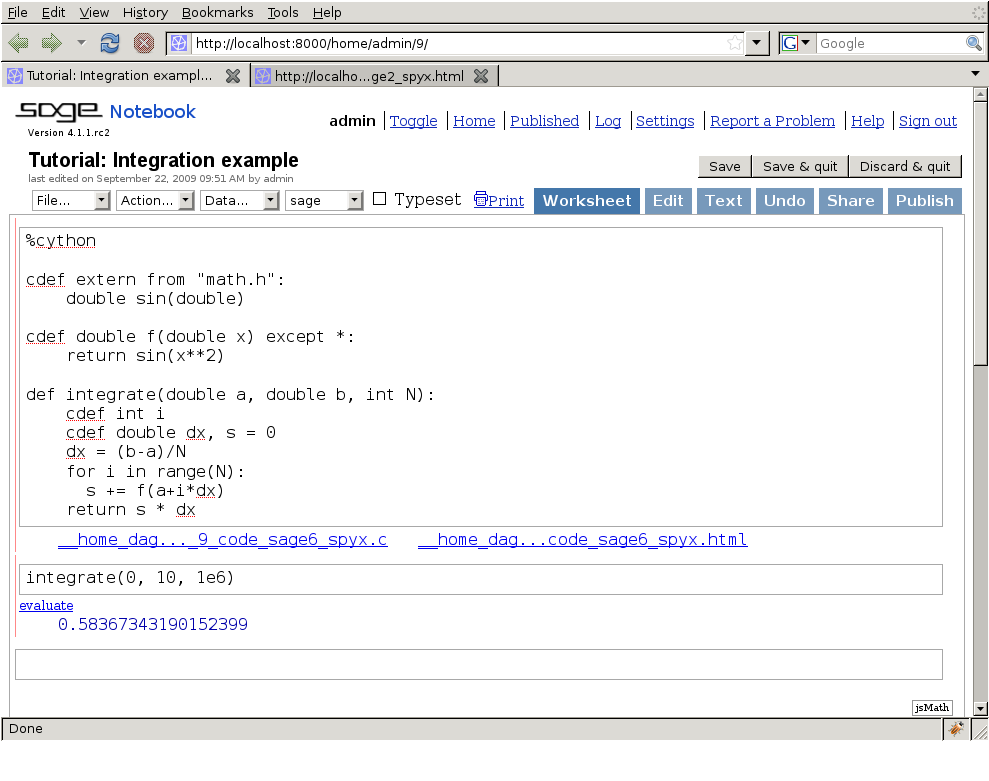
For users of the Sage math distribution, the Sage notebook allows transparently editing and compiling Cython code simply by typing %cython at the top of a cell and evaluate it. Variables and functions defined in a Cython cell imported into the running session.
| [IPython] | http://ipython.org |
| [Sage] |
|
