Note
Click here to download the full example code
Classification of text documents using sparse features¶
This is an example showing how scikit-learn can be used to classify documents by topics using a bag-of-words approach. This example uses a scipy.sparse matrix to store the features and demonstrates various classifiers that can efficiently handle sparse matrices.
The dataset used in this example is the 20 newsgroups dataset. It will be automatically downloaded, then cached.
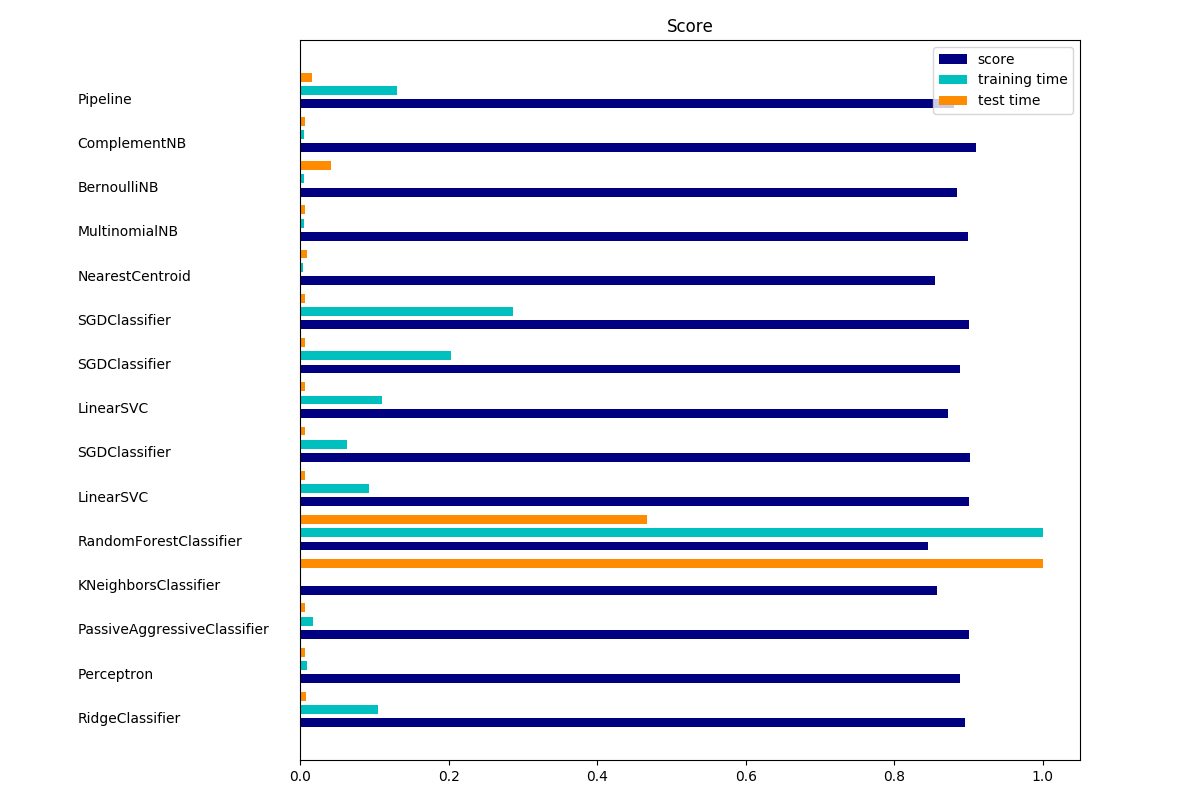
The bar plot indicates the accuracy, training time (normalized) and test time (normalized) of each classifier.
Out:
Usage: plot_document_classification_20newsgroups.py [options]
Options:
-h, --help show this help message and exit
--report Print a detailed classification report.
--chi2_select=SELECT_CHI2
Select some number of features using a chi-squared
test
--confusion_matrix Print the confusion matrix.
--top10 Print ten most discriminative terms per class for
every classifier.
--all_categories Whether to use all categories or not.
--use_hashing Use a hashing vectorizer.
--n_features=N_FEATURES
n_features when using the hashing vectorizer.
--filtered Remove newsgroup information that is easily overfit:
headers, signatures, and quoting.
Loading 20 newsgroups dataset for categories:
['alt.atheism', 'talk.religion.misc', 'comp.graphics', 'sci.space']
data loaded
2034 documents - 3.980MB (training set)
1353 documents - 2.867MB (test set)
4 categories
Extracting features from the training data using a sparse vectorizer
done in 0.718310s at 5.540MB/s
n_samples: 2034, n_features: 33809
Extracting features from the test data using the same vectorizer
done in 0.459944s at 6.234MB/s
n_samples: 1353, n_features: 33809
================================================================================
Ridge Classifier
________________________________________________________________________________
Training:
RidgeClassifier(alpha=1.0, class_weight=None, copy_X=True, fit_intercept=True,
max_iter=None, normalize=False, random_state=None, solver='sag',
tol=0.01)
train time: 0.205s
test time: 0.002s
accuracy: 0.896
dimensionality: 33809
density: 1.000000
================================================================================
Perceptron
________________________________________________________________________________
Training:
Perceptron(alpha=0.0001, class_weight=None, early_stopping=False, eta0=1.0,
fit_intercept=True, max_iter=50, n_iter=None, n_iter_no_change=5,
n_jobs=None, penalty=None, random_state=0, shuffle=True, tol=0.001,
validation_fraction=0.1, verbose=0, warm_start=False)
train time: 0.019s
test time: 0.002s
accuracy: 0.888
dimensionality: 33809
density: 0.240114
================================================================================
Passive-Aggressive
________________________________________________________________________________
Training:
PassiveAggressiveClassifier(C=1.0, average=False, class_weight=None,
early_stopping=False, fit_intercept=True, loss='hinge',
max_iter=50, n_iter=None, n_iter_no_change=5, n_jobs=None,
random_state=None, shuffle=True, tol=0.001,
validation_fraction=0.1, verbose=0, warm_start=False)
train time: 0.033s
test time: 0.002s
accuracy: 0.900
dimensionality: 33809
density: 0.702069
================================================================================
kNN
________________________________________________________________________________
Training:
KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski',
metric_params=None, n_jobs=None, n_neighbors=10, p=2,
weights='uniform')
train time: 0.002s
test time: 0.247s
accuracy: 0.858
================================================================================
Random forest
________________________________________________________________________________
Training:
RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini',
max_depth=None, max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=100, n_jobs=None,
oob_score=False, random_state=None, verbose=0,
warm_start=False)
train time: 1.937s
test time: 0.115s
accuracy: 0.845
================================================================================
L2 penalty
________________________________________________________________________________
Training:
LinearSVC(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, loss='squared_hinge', max_iter=1000,
multi_class='ovr', penalty='l2', random_state=None, tol=0.001,
verbose=0)
train time: 0.179s
test time: 0.002s
accuracy: 0.900
dimensionality: 33809
density: 1.000000
________________________________________________________________________________
Training:
SGDClassifier(alpha=0.0001, average=False, class_weight=None,
early_stopping=False, epsilon=0.1, eta0=0.0, fit_intercept=True,
l1_ratio=0.15, learning_rate='optimal', loss='hinge', max_iter=50,
n_iter=None, n_iter_no_change=5, n_jobs=None, penalty='l2',
power_t=0.5, random_state=None, shuffle=True, tol=None,
validation_fraction=0.1, verbose=0, warm_start=False)
train time: 0.121s
test time: 0.002s
accuracy: 0.902
dimensionality: 33809
density: 0.666213
================================================================================
L1 penalty
________________________________________________________________________________
Training:
LinearSVC(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, loss='squared_hinge', max_iter=1000,
multi_class='ovr', penalty='l1', random_state=None, tol=0.001,
verbose=0)
train time: 0.213s
test time: 0.002s
accuracy: 0.873
dimensionality: 33809
density: 0.005575
________________________________________________________________________________
Training:
SGDClassifier(alpha=0.0001, average=False, class_weight=None,
early_stopping=False, epsilon=0.1, eta0=0.0, fit_intercept=True,
l1_ratio=0.15, learning_rate='optimal', loss='hinge', max_iter=50,
n_iter=None, n_iter_no_change=5, n_jobs=None, penalty='l1',
power_t=0.5, random_state=None, shuffle=True, tol=None,
validation_fraction=0.1, verbose=0, warm_start=False)
train time: 0.393s
test time: 0.002s
accuracy: 0.888
dimensionality: 33809
density: 0.020128
================================================================================
Elastic-Net penalty
________________________________________________________________________________
Training:
SGDClassifier(alpha=0.0001, average=False, class_weight=None,
early_stopping=False, epsilon=0.1, eta0=0.0, fit_intercept=True,
l1_ratio=0.15, learning_rate='optimal', loss='hinge', max_iter=50,
n_iter=None, n_iter_no_change=5, n_jobs=None, penalty='elasticnet',
power_t=0.5, random_state=None, shuffle=True, tol=None,
validation_fraction=0.1, verbose=0, warm_start=False)
train time: 0.555s
test time: 0.002s
accuracy: 0.901
dimensionality: 33809
density: 0.186615
================================================================================
NearestCentroid (aka Rocchio classifier)
________________________________________________________________________________
Training:
NearestCentroid(metric='euclidean', shrink_threshold=None)
train time: 0.009s
test time: 0.002s
accuracy: 0.855
================================================================================
Naive Bayes
________________________________________________________________________________
Training:
MultinomialNB(alpha=0.01, class_prior=None, fit_prior=True)
train time: 0.009s
test time: 0.002s
accuracy: 0.899
dimensionality: 33809
density: 1.000000
________________________________________________________________________________
Training:
BernoulliNB(alpha=0.01, binarize=0.0, class_prior=None, fit_prior=True)
train time: 0.011s
test time: 0.010s
accuracy: 0.884
dimensionality: 33809
density: 1.000000
________________________________________________________________________________
Training:
ComplementNB(alpha=0.1, class_prior=None, fit_prior=True, norm=False)
train time: 0.011s
test time: 0.002s
accuracy: 0.911
dimensionality: 33809
density: 1.000000
================================================================================
LinearSVC with L1-based feature selection
________________________________________________________________________________
Training:
Pipeline(memory=None,
steps=[('feature_selection', SelectFromModel(estimator=LinearSVC(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, loss='squared_hinge', max_iter=1000,
multi_class='ovr', penalty='l1', random_state=None, tol=0.001,
verbose=0),
max_features=None, no...ax_iter=1000,
multi_class='ovr', penalty='l2', random_state=None, tol=0.0001,
verbose=0))])
train time: 0.254s
test time: 0.004s
accuracy: 0.880
# Author: Peter Prettenhofer <peter.prettenhofer@gmail.com>
# Olivier Grisel <olivier.grisel@ensta.org>
# Mathieu Blondel <mathieu@mblondel.org>
# Lars Buitinck
# License: BSD 3 clause
from __future__ import print_function
import logging
import numpy as np
from optparse import OptionParser
import sys
from time import time
import matplotlib.pyplot as plt
from sklearn.datasets import fetch_20newsgroups
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.feature_extraction.text import HashingVectorizer
from sklearn.feature_selection import SelectFromModel
from sklearn.feature_selection import SelectKBest, chi2
from sklearn.linear_model import RidgeClassifier
from sklearn.pipeline import Pipeline
from sklearn.svm import LinearSVC
from sklearn.linear_model import SGDClassifier
from sklearn.linear_model import Perceptron
from sklearn.linear_model import PassiveAggressiveClassifier
from sklearn.naive_bayes import BernoulliNB, ComplementNB, MultinomialNB
from sklearn.neighbors import KNeighborsClassifier
from sklearn.neighbors import NearestCentroid
from sklearn.ensemble import RandomForestClassifier
from sklearn.utils.extmath import density
from sklearn import metrics
# Display progress logs on stdout
logging.basicConfig(level=logging.INFO,
format='%(asctime)s %(levelname)s %(message)s')
# parse commandline arguments
op = OptionParser()
op.add_option("--report",
action="store_true", dest="print_report",
help="Print a detailed classification report.")
op.add_option("--chi2_select",
action="store", type="int", dest="select_chi2",
help="Select some number of features using a chi-squared test")
op.add_option("--confusion_matrix",
action="store_true", dest="print_cm",
help="Print the confusion matrix.")
op.add_option("--top10",
action="store_true", dest="print_top10",
help="Print ten most discriminative terms per class"
" for every classifier.")
op.add_option("--all_categories",
action="store_true", dest="all_categories",
help="Whether to use all categories or not.")
op.add_option("--use_hashing",
action="store_true",
help="Use a hashing vectorizer.")
op.add_option("--n_features",
action="store", type=int, default=2 ** 16,
help="n_features when using the hashing vectorizer.")
op.add_option("--filtered",
action="store_true",
help="Remove newsgroup information that is easily overfit: "
"headers, signatures, and quoting.")
def is_interactive():
return not hasattr(sys.modules['__main__'], '__file__')
# work-around for Jupyter notebook and IPython console
argv = [] if is_interactive() else sys.argv[1:]
(opts, args) = op.parse_args(argv)
if len(args) > 0:
op.error("this script takes no arguments.")
sys.exit(1)
print(__doc__)
op.print_help()
print()
# #############################################################################
# Load some categories from the training set
if opts.all_categories:
categories = None
else:
categories = [
'alt.atheism',
'talk.religion.misc',
'comp.graphics',
'sci.space',
]
if opts.filtered:
remove = ('headers', 'footers', 'quotes')
else:
remove = ()
print("Loading 20 newsgroups dataset for categories:")
print(categories if categories else "all")
data_train = fetch_20newsgroups(subset='train', categories=categories,
shuffle=True, random_state=42,
remove=remove)
data_test = fetch_20newsgroups(subset='test', categories=categories,
shuffle=True, random_state=42,
remove=remove)
print('data loaded')
# order of labels in `target_names` can be different from `categories`
target_names = data_train.target_names
def size_mb(docs):
return sum(len(s.encode('utf-8')) for s in docs) / 1e6
data_train_size_mb = size_mb(data_train.data)
data_test_size_mb = size_mb(data_test.data)
print("%d documents - %0.3fMB (training set)" % (
len(data_train.data), data_train_size_mb))
print("%d documents - %0.3fMB (test set)" % (
len(data_test.data), data_test_size_mb))
print("%d categories" % len(target_names))
print()
# split a training set and a test set
y_train, y_test = data_train.target, data_test.target
print("Extracting features from the training data using a sparse vectorizer")
t0 = time()
if opts.use_hashing:
vectorizer = HashingVectorizer(stop_words='english', alternate_sign=False,
n_features=opts.n_features)
X_train = vectorizer.transform(data_train.data)
else:
vectorizer = TfidfVectorizer(sublinear_tf=True, max_df=0.5,
stop_words='english')
X_train = vectorizer.fit_transform(data_train.data)
duration = time() - t0
print("done in %fs at %0.3fMB/s" % (duration, data_train_size_mb / duration))
print("n_samples: %d, n_features: %d" % X_train.shape)
print()
print("Extracting features from the test data using the same vectorizer")
t0 = time()
X_test = vectorizer.transform(data_test.data)
duration = time() - t0
print("done in %fs at %0.3fMB/s" % (duration, data_test_size_mb / duration))
print("n_samples: %d, n_features: %d" % X_test.shape)
print()
# mapping from integer feature name to original token string
if opts.use_hashing:
feature_names = None
else:
feature_names = vectorizer.get_feature_names()
if opts.select_chi2:
print("Extracting %d best features by a chi-squared test" %
opts.select_chi2)
t0 = time()
ch2 = SelectKBest(chi2, k=opts.select_chi2)
X_train = ch2.fit_transform(X_train, y_train)
X_test = ch2.transform(X_test)
if feature_names:
# keep selected feature names
feature_names = [feature_names[i] for i
in ch2.get_support(indices=True)]
print("done in %fs" % (time() - t0))
print()
if feature_names:
feature_names = np.asarray(feature_names)
def trim(s):
"""Trim string to fit on terminal (assuming 80-column display)"""
return s if len(s) <= 80 else s[:77] + "..."
# #############################################################################
# Benchmark classifiers
def benchmark(clf):
print('_' * 80)
print("Training: ")
print(clf)
t0 = time()
clf.fit(X_train, y_train)
train_time = time() - t0
print("train time: %0.3fs" % train_time)
t0 = time()
pred = clf.predict(X_test)
test_time = time() - t0
print("test time: %0.3fs" % test_time)
score = metrics.accuracy_score(y_test, pred)
print("accuracy: %0.3f" % score)
if hasattr(clf, 'coef_'):
print("dimensionality: %d" % clf.coef_.shape[1])
print("density: %f" % density(clf.coef_))
if opts.print_top10 and feature_names is not None:
print("top 10 keywords per class:")
for i, label in enumerate(target_names):
top10 = np.argsort(clf.coef_[i])[-10:]
print(trim("%s: %s" % (label, " ".join(feature_names[top10]))))
print()
if opts.print_report:
print("classification report:")
print(metrics.classification_report(y_test, pred,
target_names=target_names))
if opts.print_cm:
print("confusion matrix:")
print(metrics.confusion_matrix(y_test, pred))
print()
clf_descr = str(clf).split('(')[0]
return clf_descr, score, train_time, test_time
results = []
for clf, name in (
(RidgeClassifier(tol=1e-2, solver="sag"), "Ridge Classifier"),
(Perceptron(max_iter=50, tol=1e-3), "Perceptron"),
(PassiveAggressiveClassifier(max_iter=50, tol=1e-3),
"Passive-Aggressive"),
(KNeighborsClassifier(n_neighbors=10), "kNN"),
(RandomForestClassifier(n_estimators=100), "Random forest")):
print('=' * 80)
print(name)
results.append(benchmark(clf))
for penalty in ["l2", "l1"]:
print('=' * 80)
print("%s penalty" % penalty.upper())
# Train Liblinear model
results.append(benchmark(LinearSVC(penalty=penalty, dual=False,
tol=1e-3)))
# Train SGD model
results.append(benchmark(SGDClassifier(alpha=.0001, max_iter=50,
penalty=penalty)))
# Train SGD with Elastic Net penalty
print('=' * 80)
print("Elastic-Net penalty")
results.append(benchmark(SGDClassifier(alpha=.0001, max_iter=50,
penalty="elasticnet")))
# Train NearestCentroid without threshold
print('=' * 80)
print("NearestCentroid (aka Rocchio classifier)")
results.append(benchmark(NearestCentroid()))
# Train sparse Naive Bayes classifiers
print('=' * 80)
print("Naive Bayes")
results.append(benchmark(MultinomialNB(alpha=.01)))
results.append(benchmark(BernoulliNB(alpha=.01)))
results.append(benchmark(ComplementNB(alpha=.1)))
print('=' * 80)
print("LinearSVC with L1-based feature selection")
# The smaller C, the stronger the regularization.
# The more regularization, the more sparsity.
results.append(benchmark(Pipeline([
('feature_selection', SelectFromModel(LinearSVC(penalty="l1", dual=False,
tol=1e-3))),
('classification', LinearSVC(penalty="l2"))])))
# make some plots
indices = np.arange(len(results))
results = [[x[i] for x in results] for i in range(4)]
clf_names, score, training_time, test_time = results
training_time = np.array(training_time) / np.max(training_time)
test_time = np.array(test_time) / np.max(test_time)
plt.figure(figsize=(12, 8))
plt.title("Score")
plt.barh(indices, score, .2, label="score", color='navy')
plt.barh(indices + .3, training_time, .2, label="training time",
color='c')
plt.barh(indices + .6, test_time, .2, label="test time", color='darkorange')
plt.yticks(())
plt.legend(loc='best')
plt.subplots_adjust(left=.25)
plt.subplots_adjust(top=.95)
plt.subplots_adjust(bottom=.05)
for i, c in zip(indices, clf_names):
plt.text(-.3, i, c)
plt.show()
Total running time of the script: ( 0 minutes 6.271 seconds)

