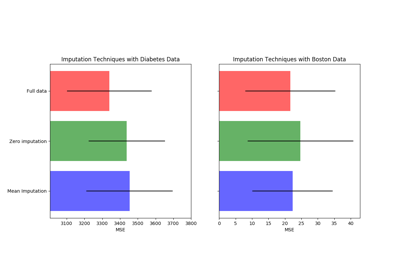
sklearn.impute.SimpleImputer¶
-
class
sklearn.impute.SimpleImputer(missing_values=nan, strategy='mean', fill_value=None, verbose=0, copy=True)[source]¶ Imputation transformer for completing missing values.
Read more in the User Guide.
Parameters: - missing_values : number, string, np.nan (default) or None
The placeholder for the missing values. All occurrences of missing_values will be imputed.
- strategy : string, optional (default=”mean”)
The imputation strategy.
- If “mean”, then replace missing values using the mean along each column. Can only be used with numeric data.
- If “median”, then replace missing values using the median along each column. Can only be used with numeric data.
- If “most_frequent”, then replace missing using the most frequent value along each column. Can be used with strings or numeric data.
- If “constant”, then replace missing values with fill_value. Can be used with strings or numeric data.
New in version 0.20: strategy=”constant” for fixed value imputation.
- fill_value : string or numerical value, optional (default=None)
When strategy == “constant”, fill_value is used to replace all occurrences of missing_values. If left to the default, fill_value will be 0 when imputing numerical data and “missing_value” for strings or object data types.
- verbose : integer, optional (default=0)
Controls the verbosity of the imputer.
- copy : boolean, optional (default=True)
If True, a copy of X will be created. If False, imputation will be done in-place whenever possible. Note that, in the following cases, a new copy will always be made, even if copy=False:
- If X is not an array of floating values;
- If X is encoded as a CSR matrix.
Attributes: - statistics_ : array of shape (n_features,)
The imputation fill value for each feature.
Notes
Columns which only contained missing values at fit are discarded upon transform if strategy is not “constant”.
Examples
>>> import numpy as np >>> from sklearn.impute import SimpleImputer >>> imp_mean = SimpleImputer(missing_values=np.nan, strategy='mean') >>> imp_mean.fit([[7, 2, 3], [4, np.nan, 6], [10, 5, 9]]) ... # doctest: +NORMALIZE_WHITESPACE SimpleImputer(copy=True, fill_value=None, missing_values=nan, strategy='mean', verbose=0) >>> X = [[np.nan, 2, 3], [4, np.nan, 6], [10, np.nan, 9]] >>> print(imp_mean.transform(X)) ... # doctest: +NORMALIZE_WHITESPACE [[ 7. 2. 3. ] [ 4. 3.5 6. ] [10. 3.5 9. ]]
Methods
fit(X[, y])Fit the imputer on X. fit_transform(X[, y])Fit to data, then transform it. get_params([deep])Get parameters for this estimator. set_params(**params)Set the parameters of this estimator. transform(X)Impute all missing values in X. -
__init__(missing_values=nan, strategy='mean', fill_value=None, verbose=0, copy=True)[source]¶ Initialize self. See help(type(self)) for accurate signature.
-
fit(X, y=None)[source]¶ Fit the imputer on X.
Parameters: - X : {array-like, sparse matrix}, shape (n_samples, n_features)
Input data, where
n_samplesis the number of samples andn_featuresis the number of features.
Returns: - self : SimpleImputer
-
fit_transform(X, y=None, **fit_params)[source]¶ Fit to data, then transform it.
Fits transformer to X and y with optional parameters fit_params and returns a transformed version of X.
Parameters: - X : numpy array of shape [n_samples, n_features]
Training set.
- y : numpy array of shape [n_samples]
Target values.
Returns: - X_new : numpy array of shape [n_samples, n_features_new]
Transformed array.
-
get_params(deep=True)[source]¶ Get parameters for this estimator.
Parameters: - deep : boolean, optional
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: - params : mapping of string to any
Parameter names mapped to their values.
-
set_params(**params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.Returns: - self