Printing/Drawing Theano graphs¶
Theano provides the functions theano.printing.pprint() and
theano.printing.debugprint() to print a graph to the terminal before or
after compilation. pprint() is more compact and math-like,
debugprint() is more verbose. Theano also provides pydotprint()
that creates an image of the function. You can read about them in
printing – Graph Printing and Symbolic Print Statement.
Note
When printing Theano functions, they can sometimes be hard to
read. To help with this, you can disable some Theano optimizations
by using the Theano flag:
optimizer_excluding=fusion:inplace. Do not use this during
real job execution, as this will make the graph slower and use more
memory.
Consider again the logistic regression example:
>>> import numpy
>>> import theano
>>> import theano.tensor as T
>>> rng = numpy.random
>>> # Training data
>>> N = 400
>>> feats = 784
>>> D = (rng.randn(N, feats).astype(theano.config.floatX), rng.randint(size=N,low=0, high=2).astype(theano.config.floatX))
>>> training_steps = 10000
>>> # Declare Theano symbolic variables
>>> x = T.matrix("x")
>>> y = T.vector("y")
>>> w = theano.shared(rng.randn(feats).astype(theano.config.floatX), name="w")
>>> b = theano.shared(numpy.asarray(0., dtype=theano.config.floatX), name="b")
>>> x.tag.test_value = D[0]
>>> y.tag.test_value = D[1]
>>> # Construct Theano expression graph
>>> p_1 = 1 / (1 + T.exp(-T.dot(x, w)-b)) # Probability of having a one
>>> prediction = p_1 > 0.5 # The prediction that is done: 0 or 1
>>> # Compute gradients
>>> xent = -y*T.log(p_1) - (1-y)*T.log(1-p_1) # Cross-entropy
>>> cost = xent.mean() + 0.01*(w**2).sum() # The cost to optimize
>>> gw,gb = T.grad(cost, [w,b])
>>> # Training and prediction function
>>> train = theano.function(inputs=[x,y], outputs=[prediction, xent], updates=[[w, w-0.01*gw], [b, b-0.01*gb]], name = "train")
>>> predict = theano.function(inputs=[x], outputs=prediction, name = "predict")
Pretty Printing¶
>>> theano.printing.pprint(prediction)
'gt((TensorConstant{1} / (TensorConstant{1} + exp(((-(x \\dot w)) - b)))),
TensorConstant{0.5})'
Debug Print¶
The pre-compilation graph:
>>> theano.printing.debugprint(prediction)
Elemwise{gt,no_inplace} [id A] ''
|Elemwise{true_div,no_inplace} [id B] ''
| |DimShuffle{x} [id C] ''
| | |TensorConstant{1} [id D]
| |Elemwise{add,no_inplace} [id E] ''
| |DimShuffle{x} [id F] ''
| | |TensorConstant{1} [id D]
| |Elemwise{exp,no_inplace} [id G] ''
| |Elemwise{sub,no_inplace} [id H] ''
| |Elemwise{neg,no_inplace} [id I] ''
| | |dot [id J] ''
| | |x [id K]
| | |w [id L]
| |DimShuffle{x} [id M] ''
| |b [id N]
|DimShuffle{x} [id O] ''
|TensorConstant{0.5} [id P]
The post-compilation graph:
>>> theano.printing.debugprint(predict)
Elemwise{Composite{GT(scalar_sigmoid((-((-i0) - i1))), i2)}} [id A] '' 4
|CGemv{inplace} [id B] '' 3
| |AllocEmpty{dtype='float64'} [id C] '' 2
| | |Shape_i{0} [id D] '' 1
| | |x [id E]
| |TensorConstant{1.0} [id F]
| |x [id E]
| |w [id G]
| |TensorConstant{0.0} [id H]
|InplaceDimShuffle{x} [id I] '' 0
| |b [id J]
|TensorConstant{(1,) of 0.5} [id K]
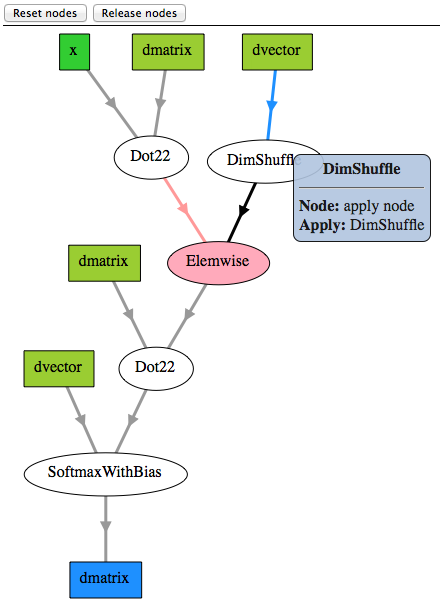
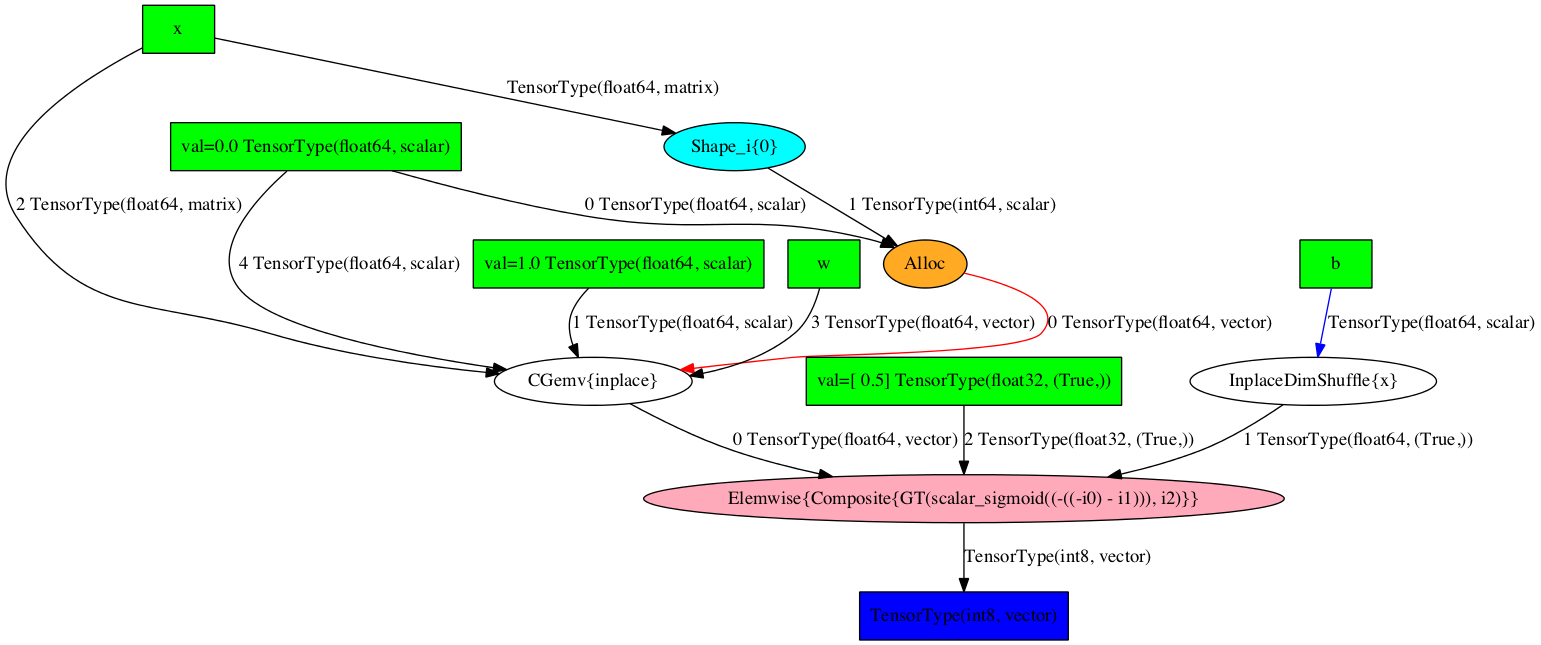
Picture Printing of Graphs¶
The pre-compilation graph:
>>> theano.printing.pydotprint(prediction, outfile="pics/logreg_pydotprint_prediction.png", var_with_name_simple=True)
The output file is available at pics/logreg_pydotprint_prediction.png

The post-compilation graph:
>>> theano.printing.pydotprint(predict, outfile="pics/logreg_pydotprint_predict.png", var_with_name_simple=True)
The output file is available at pics/logreg_pydotprint_predict.png
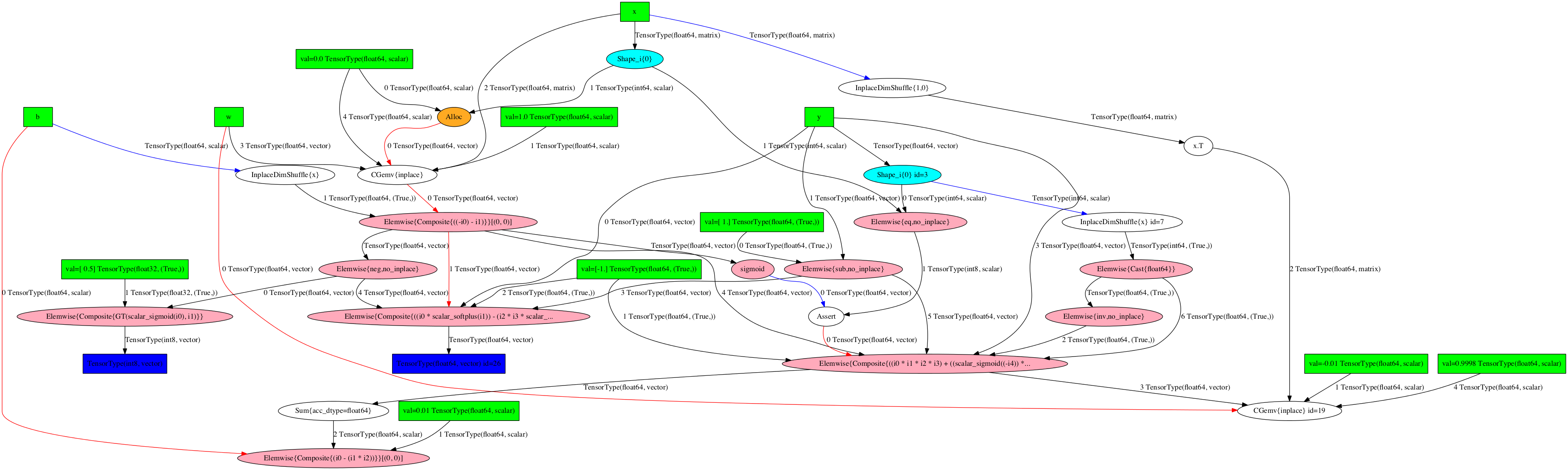
The optimized training graph:
>>> theano.printing.pydotprint(train, outfile="pics/logreg_pydotprint_train.png", var_with_name_simple=True)
The output file is available at pics/logreg_pydotprint_train.png
Interactive Graph Visualization¶
The new d3viz module complements theano.printing.pydotprint() to
visualize complex graph structures. Instead of creating a static image, it
generates an HTML file, which allows to dynamically inspect graph structures in
a web browser. Features include zooming, drag-and-drop, editing node labels, or
coloring nodes by their compute time.